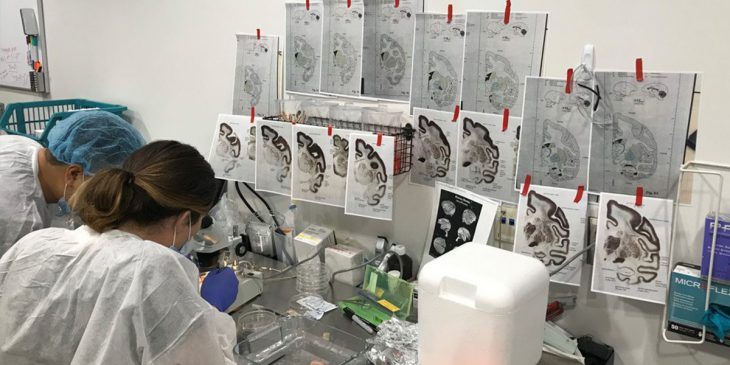
A multidisciplinary research team from the University of Pittsburgh and Carnegie Mellon University (CMU) uncovered novel insights into the molecular and spatial diversity of neurons in an area of the brain that is critical for decision making, emotions and reward.
In a recent Current Biology publication noted as Editor’s Choice in Science Magazine, researchers from the Pitt School of Medicine and CMU’s School of Computer Science identified new types of neurons in the striatum, a region of the brain involved in a variety of brain functions — from walking up stairs to deciding whether a big risk is worthwhile. The unique profiles of these neuron types provide researchers with important information on how to develop cell type-specific therapies for neurological diseases associated with abnormal function of the striatum, such as Parkinson’s disease.

Bill Stauffer, Ph.D.
“This paper is a step to much larger goals — understanding cell type-specific functions during sophisticated behaviors and establishing therapeutic tools targeting neural circuits,” said Dr. Bill Stauffer, assistant professor of neurobiology and corresponding author of the publication. “We need better ways to identify and control therapeutic targets for neurological diseases, and this work is aimed at breaking down that barrier.”
The brain is a complex system of neurons and supporting cell types, all communicating in concert to carry out the biological processes necessary for daily life. To better understand this concert of neural communication, researchers identified new players: unique types of neurons, each with potentially different connections and functions. If one type of neuron begins to play the wrong tune, this can throw off the whole symphony.
Parkinson’s disease is an example of the importance of cell type-specific functions. Deficits associated with Parkinson’s disease—slowness of movement, speech difficulties and cognitive impairments—result primarily from the loss of dopamine neurons, which severely impacts activity throughout the striatum.

Coauthors Jing He and Morgan Wirthlin perform a brain dissection
In this work, lead authors Jing He and Michael Kleyman used cutting-edge experimental tools and computational insights to characterize the diversity of medium spiny neurons, the most abundant type of neurons in the striatum. Landmark studies over the past few decades divided medium spiny neurons into two broad types, ‘D1’ and ‘D2’ neurons, that are thought to promote and suppress movements, respectively. Using new single cell technologies and machine learning, the group revealed that there are at least nine different main types of medium spiny neurons that are likely to be involved in distinct behavioral functions. With this information, scientists are now better equipped to build tools to target specific cell types and, as a result, find better treatments for the neurological diseases that they may be involved in.
Impressive projects such as the Allen Brain Atlas and the BRAIN Initiative, have set their sights on related goals. However, a big part of this and other neuroscience research is done using samples from rodents and, to a lesser extent, human samples. Mice and rats have substantial differences in gene expression and brain anatomy compared to humans, and tissue integrity of human samples is never perfect. For these reasons, Stauffer and his team chose to conduct their study on non-human primates, maximizing the likelihood that their findings can be translated into future therapies.
“One of the coolest things about this project is that it is larger in scope than anything my lab could accomplish on its own,” says Dr. Stauffer. “We normally do experiments studying reward and decision making. This project enabled us to collaborate with Andreas Pfenning from CMU and Leah Byrne from Pitt’s Department of Opthalmology, experts in computational biology and gene therapy and tackle big problems. The interdisciplinary collaboration is also what makes this work highly significant and impactful.”
Susannah Waxman is a Ph.D. candidate in the Cellular and Molecular Pathology Graduate Program at the University of Pittsburgh School of Medicine. She is participating in the UPMC Science Writing Mentorship Program